Training a classifier¶
This page illustrates how to use the HistomicsML interfaces to train classification rules using the example data provided in the Docker container.
Initializing the classifier¶
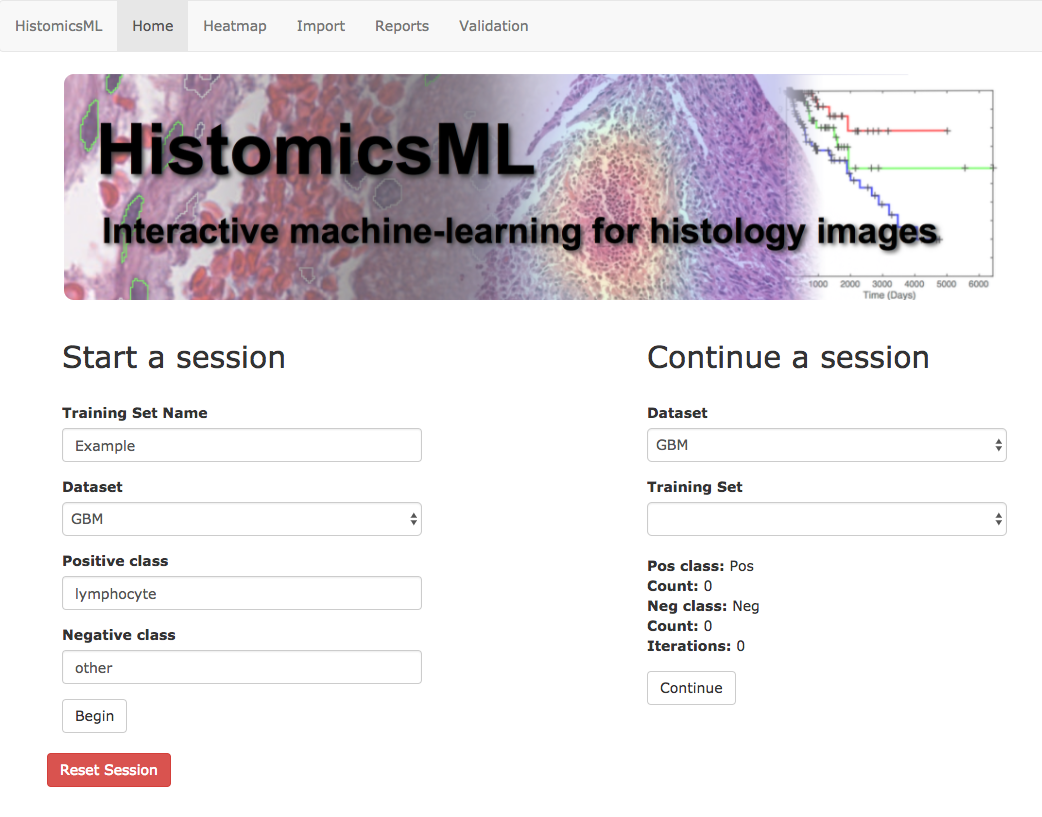
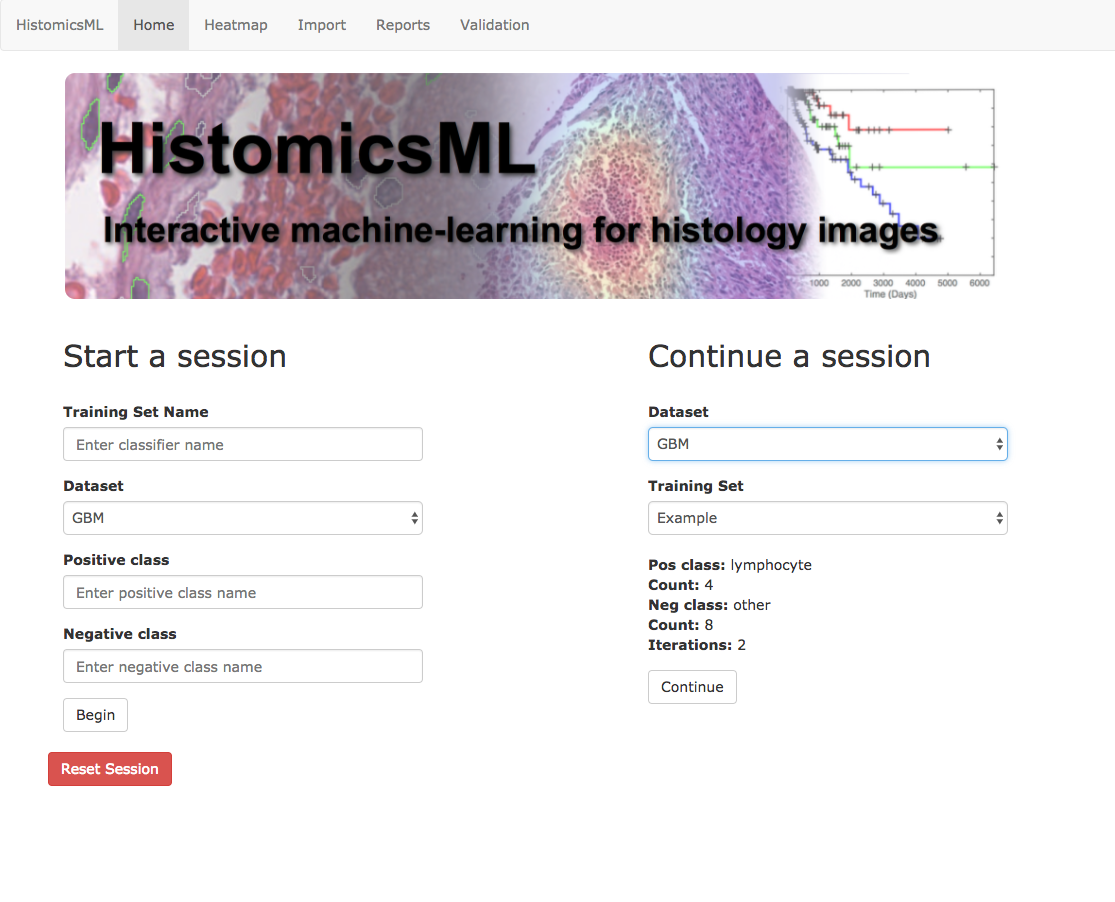
Go to http://localhost/HistomicsML/.
Under Start a session enter a training set name and select the pre-loaded glioblastoma dataset from the drop-down menu. Enter names for your classes - in our case we will use lymphocyte for the positive class and other for the negative class.
After clicking Begin the Prime interface will be loaded to collect annotations in order to initialize the classifier. The drop-down can be used to select a slides to display in the slide viewer. Zoom to a region of interest in the slide, then click Show Segmentation to display object boundaries. After clicking Select Nuclei, you will be prompted to select four objects from each class. Double-clicking an object in the slide viewer will add this object to the training set, and display a thumbnail image of the object above the slide viewer.
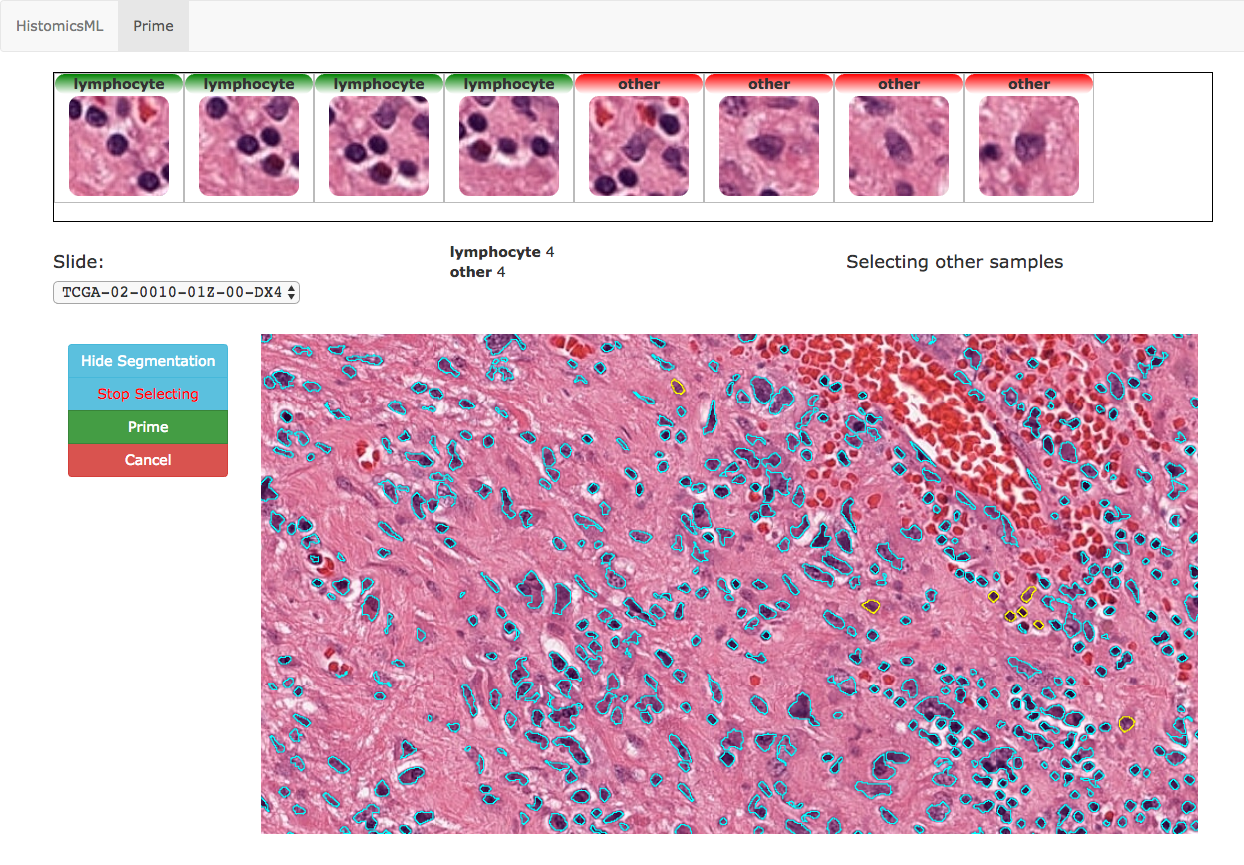
We selected 4 examples of mononuclear cells and 4 examples of other cell types in this example.
Note
You can remove objects from the training set in this menu by double clicking their thumbnail images.
With the initial annotation complete, click Prime to create the classifier. There will be a small delay while the classifier is trained and applied to the entire dataset to generate predictions for active learning.
Instance-based active learning¶
The instance-based learning interface presents objects selected by the active learning algorithm to the user for annotation. The selected objects are displayed along with their predicted class labels at the top of the slide viewer. Clicking one of these objects will bring it into focus in the slide viewer for inspection. Feedback is provided by double-clicking the thumbnails with incorrect predictions to cycle their class labels. After correcting the predictions the classifier can be updated with these new annotations by clicking Update.
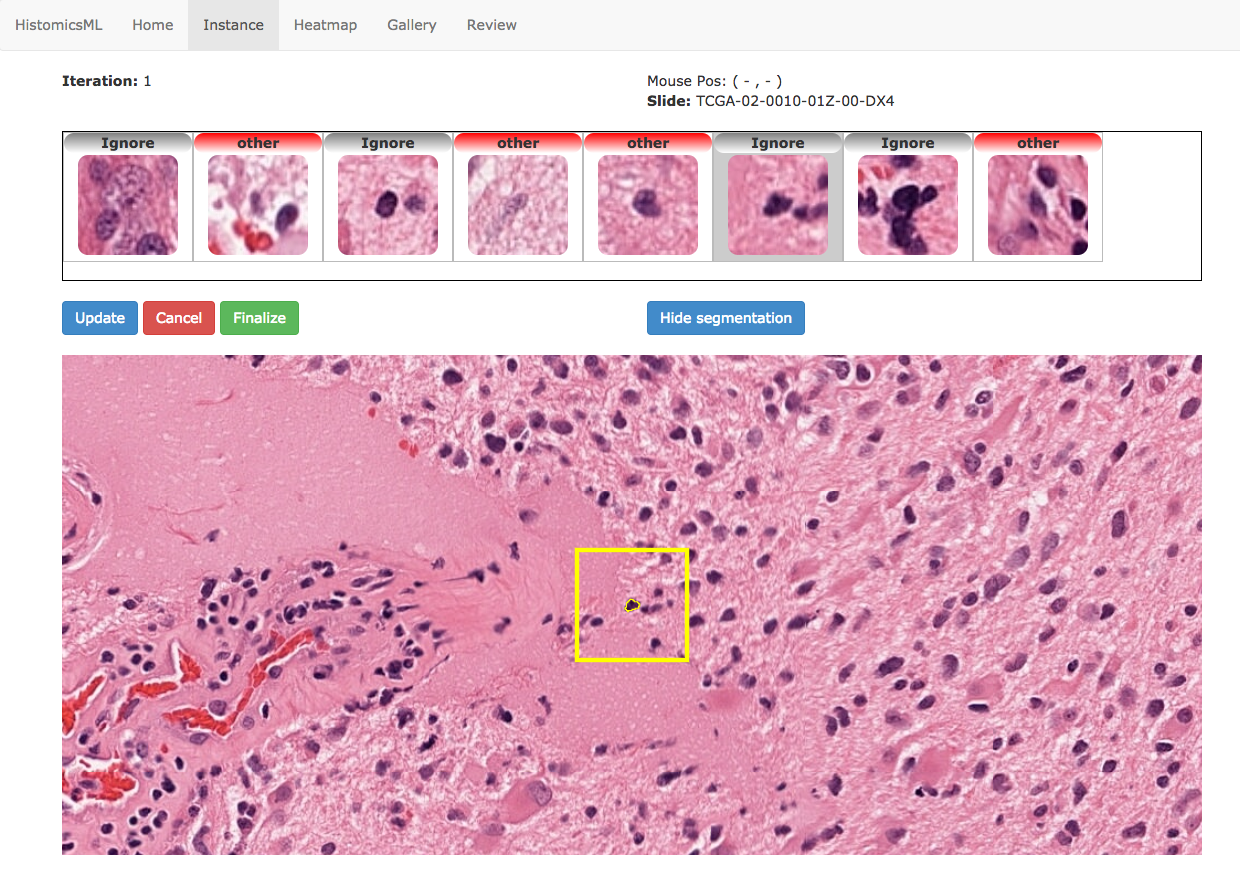
Note
Objects can be ignored in instance-based learning by assigning the ignore label. This is useful for dealing with imaging artifacts, improperly segmented objects, or objects with an ambiguous appearance that cannot be clearly labeled.
Heatmap-based active learning¶
The Gallery menu provides a high-level overview of the current classification results for the entire dataset. Each row displays heatmaps for a single slide - the left heatmap indicates the classifier uncertainty (red = more uncertain) - and the right heatmap indicates the positive class object density (red = higher density of positively classified objects). Slides are sorted in this view based on average uncertainty, with the slide having the most uncertaintly placed at the top.
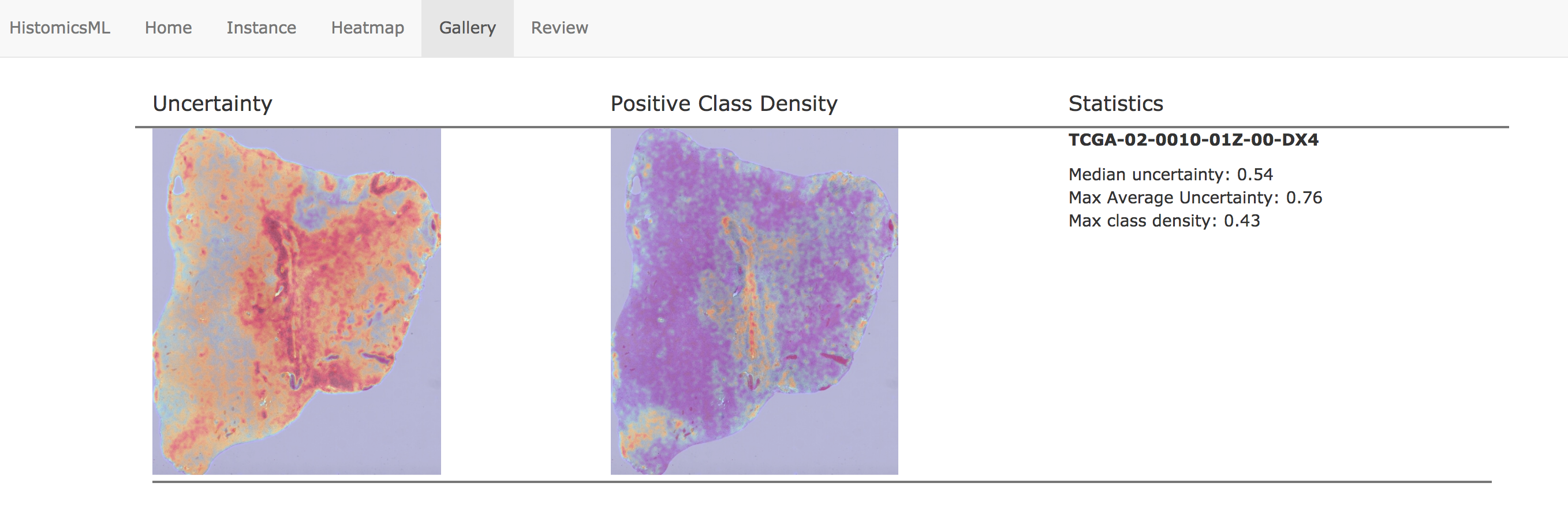
Clicking a slide in the gallery will load this slide in the heatmap viewer, where the user can identify regions for annotation. Clicking Show Segmentation will display the heatmap overlay on the slide viewer, and the user can zoom to hotspots to provide corrections to the classifier.
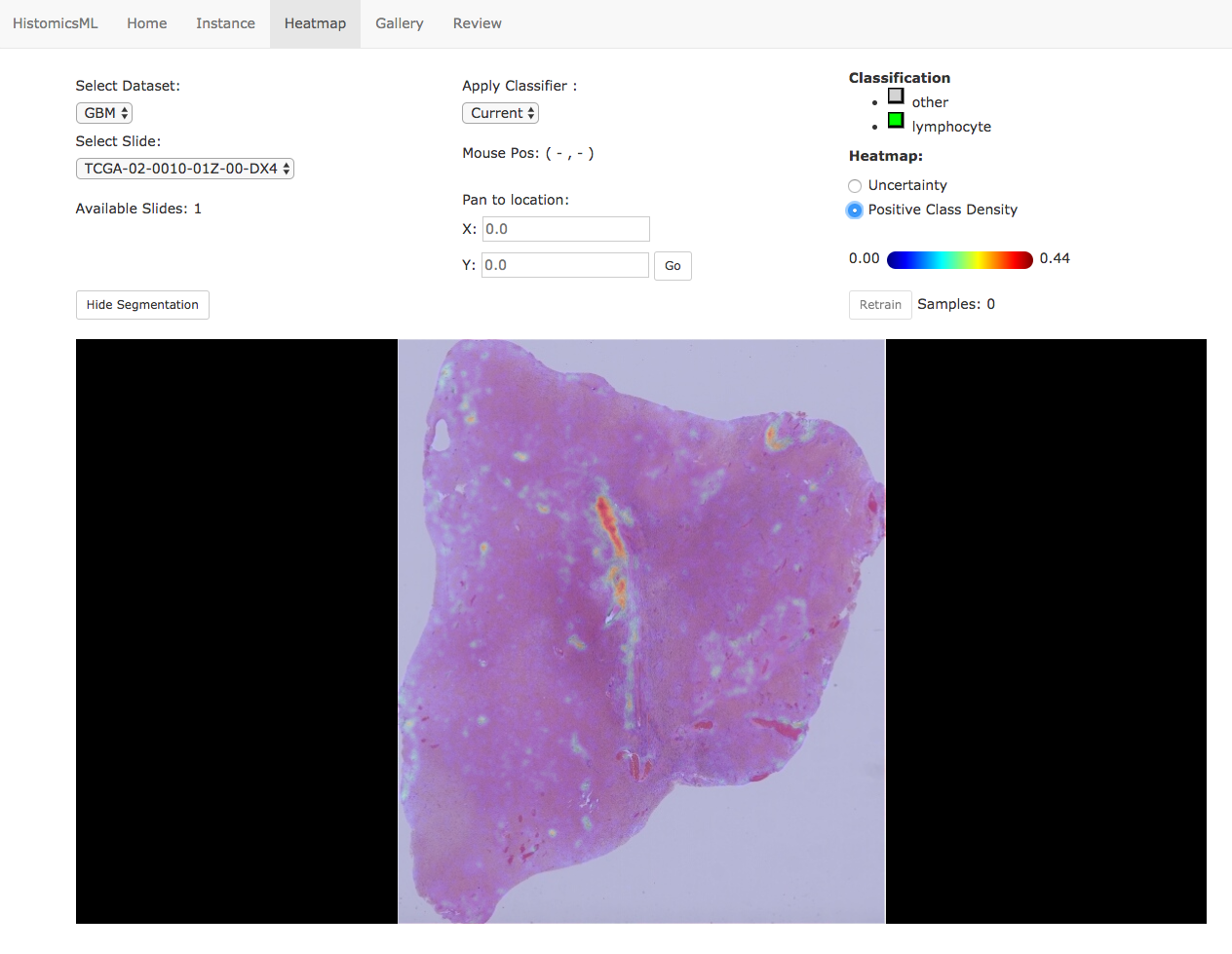
At high-magnification, objects are displayed with color-coded boundaries to indicate their predicted class (green = positive). Prediction errors can be corrected directly in the slide viewer by double clicking an object to cycle it’s class, adding this object to the training set. The classifier can be retrained with the Retrain button.
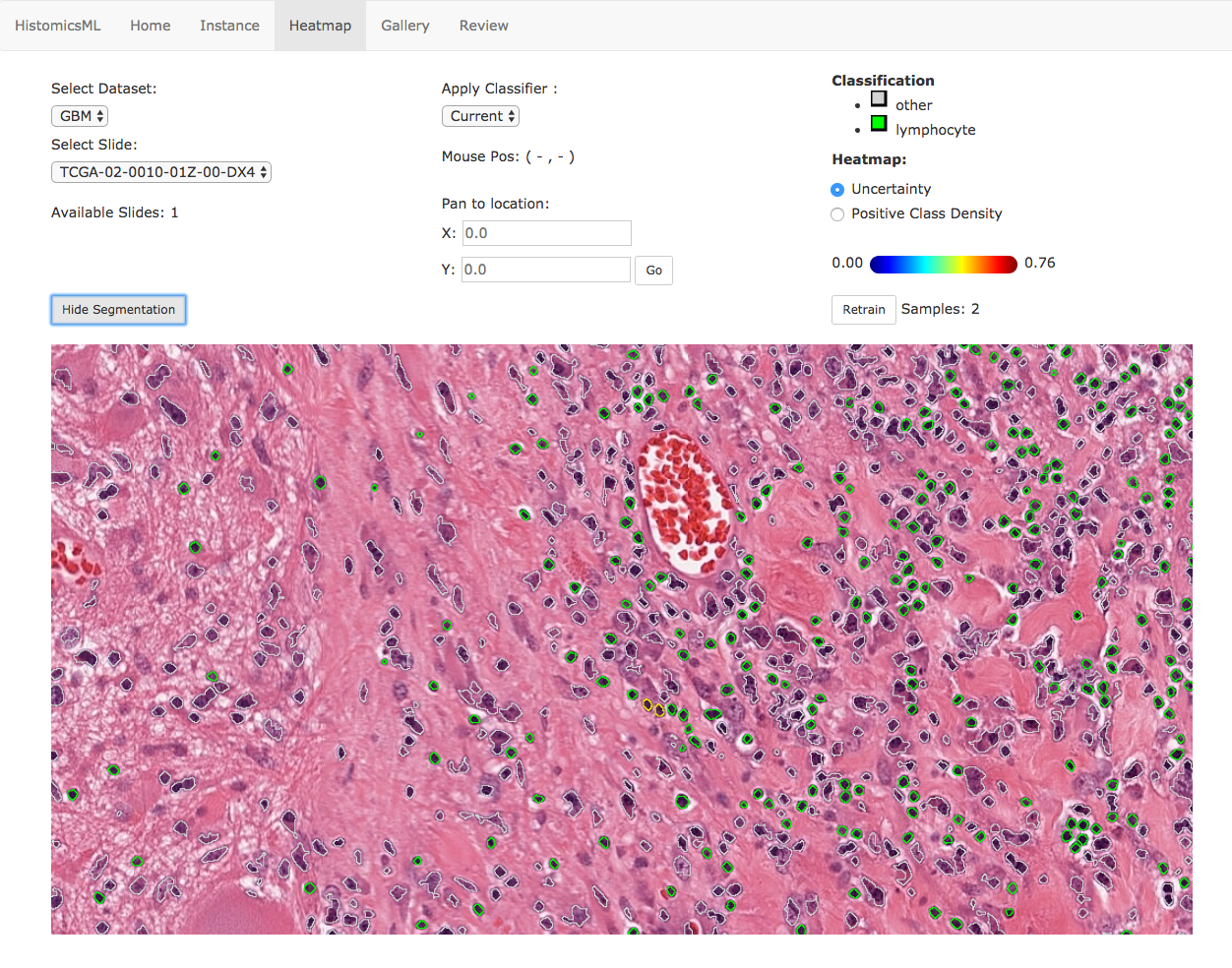
Note
Object labels can be cycled in the heatmap menu by double-click. Objects that have been annotated and added to the training set will appear with yellow boundaries. An object can be removed from the training set by double-clicking a second time.
When the training is completed, click the Finalize button in the Instance interface to save the training set to the database. This training set can be reloaded and resumed from using the Continue a session option on the main page.
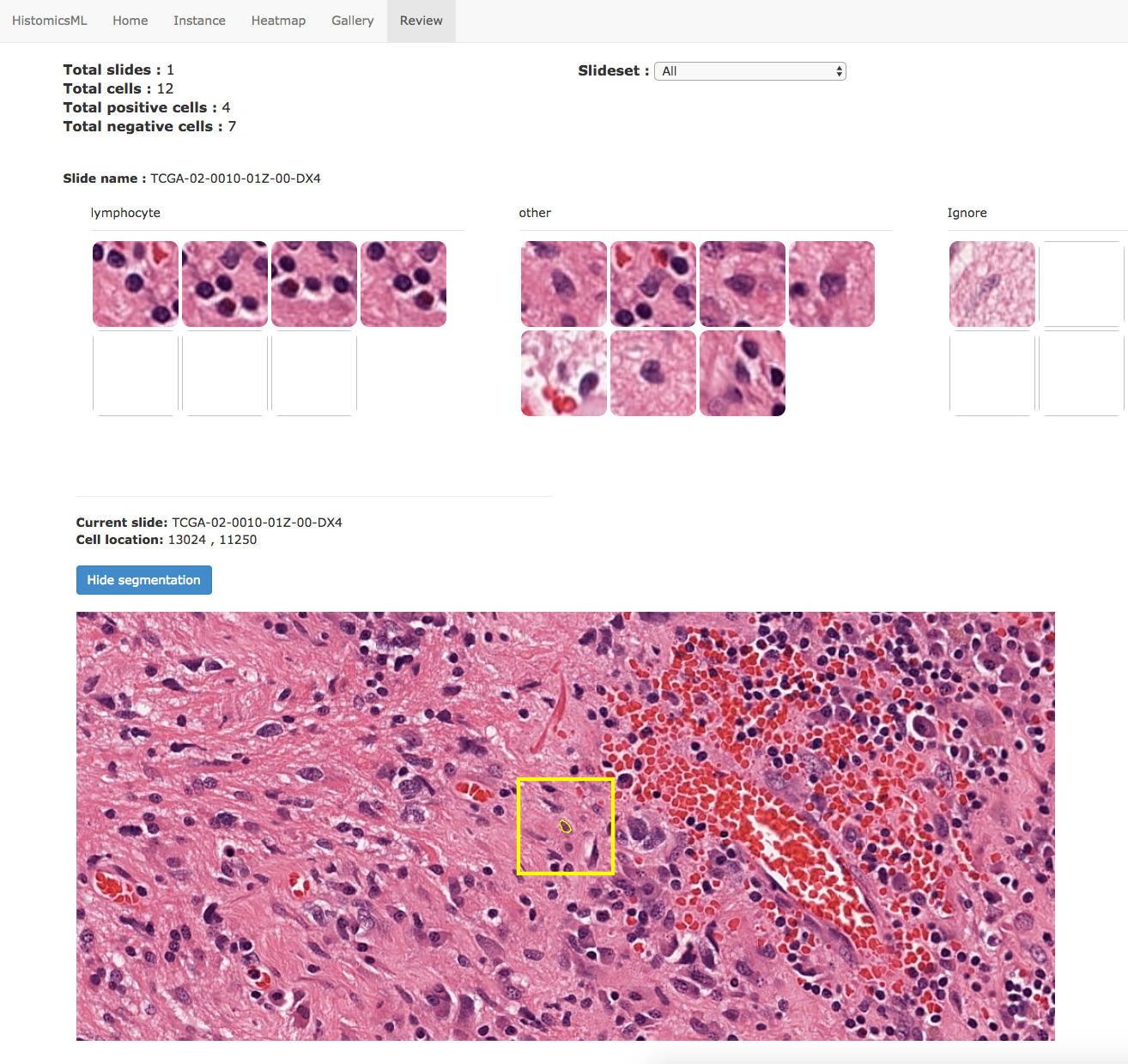
Reviewing a training set¶
Annotations in a validation set can be reviewed using the review interface.
At the home page under Continue a session, select the dataset and training set name and click Continue. Navigate to the Review interface by clicking the tab at the top menu.
The review interface displays the annotated objects organized by class and slide. Thumbnail images of the objects are organized into columns by class. Clicking a thumbnail will bring that object into the field of view in the slide view. The thumbnails can be dragged/dropped to a different column to change the class label, or placed in the Ignore column to discard them from the set. Changes are instantly commited to the database (no additional button clicks are needed).